What's in my toolbox?
Tools for doing science
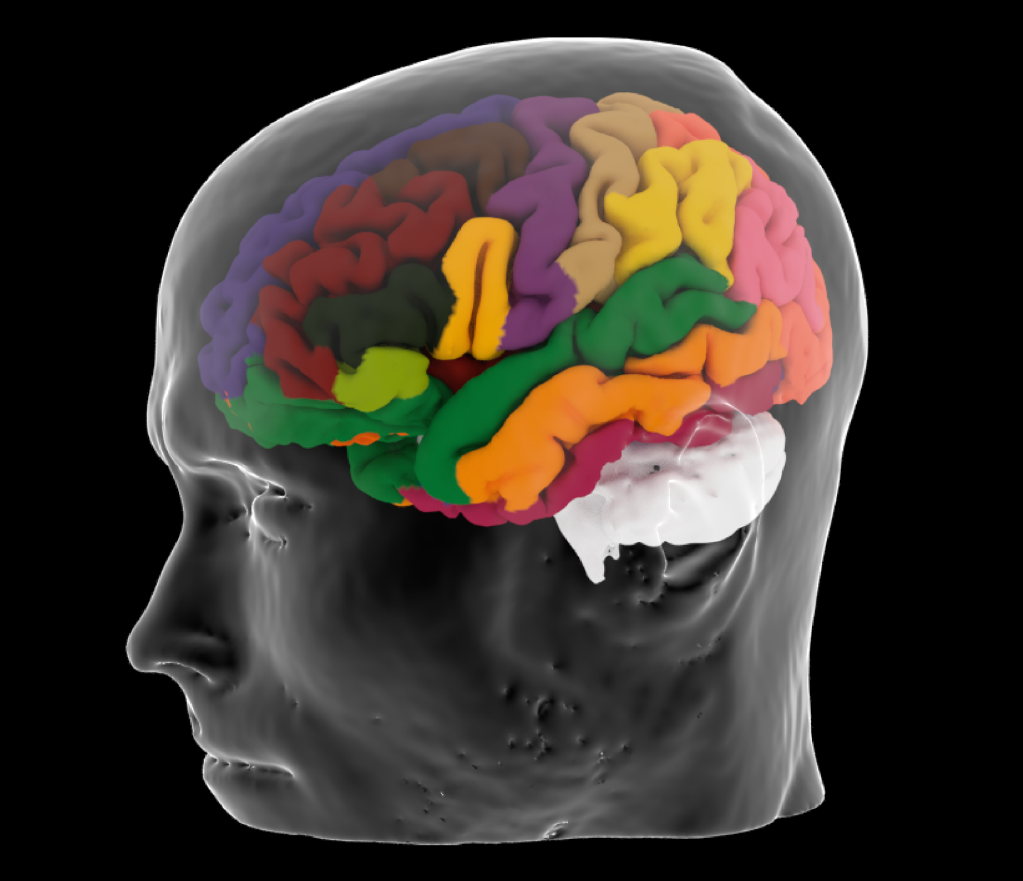
Some of the amazing visuals made possible by integrating professional grade photo- and video-editing software with state-of-the-art neuroimaging. This render from a 3T MRI scan of my brain (and head!) was done using mmvt, a freely available toolbox developed by friend and colleague, Noam Peled at the Martinos Center. Make sure to check out his work!
STABLE DIFFUSION
Stable Diffusion is an open-sourced text-to-image generator (example of a “Foundation Model”, all the rage these days…) released in September 2022. You can use it to generate surrogate data for neural nets to crunch on. For example, it’s astonishingly well-suited for generative “dreaming” in artificial sleep! I’ve also used it to create logos or project visuals, like this sleeping robot (left) for our sleepAI work at JHU APL. The Van Gogh squirrel (below) is just for me :)
gigantum
Gigantum is amazing. It makes reproducible, open-access science easy to do and easy to share. Run your Python analyses in a Jupyterlab notebook hosted through gigantum’s web interface. All of your environment packages and dependencies are handled by gigantum, and version control is done as a snapshot of your entire gigantum virtual machine! Prototype quickly, publish to the cloud, and access your work from any terminal, anywhere. Coupled with AWS (see Keras description below), do GPU-intensive, hardware-accelerated deep learning from something as portable and lightweight as a chromebook or tablet! I cannot recommend this service enough, folks. I’m never going back. Check it out here.
Keras w/ Tensorflow
Keras is a beautifully written Python package for Deep Learning. It takes care of the details by managing any of the more popular deep learning libraries (ex. Google’s Tensorflow, Torch, Theano, etc.) on the backend, so you can focus on the big picture! It’s incredibly well documented, too. Check it out here. Pro tip: I use a (free!) account with Amazon Web Services (AWS) to deploy all my neural network setups. With a p2.xlarge instance, you have access to gigabytes of free SSD storage and a K80 NVIDIA Tesla GPU with CUDA for lightning fast network training. Train networks in minutes or hours instead days!
EDF Browser
A lightweight, open-source EDFbrowser developed by Teunis van Beelen. Source code and compiled binaries are available here. The instructions on Teunis' site walk a novice through the steps to compile the app from source on Linux, macOS, and Windows. For those looking for a precompiled version to run on macOS (not available from the EDFbrowser site), you can download it here.
Neuralact
Developed by friend and colleague Jan Kubabek, NeuralAct allows researchers to visualize cortical activity acquired using ECoG, EEG, MEG, or DOT on a 3D model of the cortex. I use this extensively in my work (thanks, Jan!).
The Matlab package is available here. It's thoroughly documented and includes a nice demo.
Freesurfer
Developed by own own at the MGH Martinos Center, freesurfer is an open source software suite for processing and analyzing (human) brain MRI images. Most of my image processing chains start with freesurfer to extract and reconstruct anatomical features, which I then move to other tools for postprocessing or co-registration with other imaging modalities (ex. CT for coregistration of intracranial electrodes in SPM). A must-have for any neuroimager's toolbox!
SPM
Another must-have, SPM (Statistical Parametric Mapping) was developed for the analysis of structural and functional MRI by the Wellcome Trust Centre for Neuroimaging. SPM can also extract anatomical features, but uses a different approach than freesurfer. I think I'll post an article on the pros and cons of each sometime soon (neither is better than the other -- you just need to pick the right tool for the job, depending on your objectives). I still use SPM for my iEEG/ECoG analyses, for co-registering postoperative CT scans with preoperative MRI to localize electrode implants onto subject-specific anatomical models. And if you're an fMRI person, this is your tool, folks.
Osirix
Osirix is a great tool for viewing raw neuroimaging data (ex. DICOM, NIfTI). I use it for performing quick quality checks on my imaging data. The professional license is expensive, but there is a free license available as well, for macOS, Windows, and Linux.
GIMP
OK, so you've preprocessed your images using freesurfer, SPM, Neuralact, etc. How do you touch them up to look pro? GIMP is one tool. It's basically an open-source Adobe Illustrator alternative. It has some great functions. I'm particularly fond of its "Color-to-alpha" function, which makes working with transparencies in your images a piece of cake. Honestly if you use it for this function alone (which I do), it's worth the download. Also incredibly powerful is its “script-fu” scripting language, which through bash shell scripting or shell calls from MATLAB scripts, allows you to integrate sophisticated post-processing directly into your primary analysis scripts.
Adobe Illustrator CS6
And speaking of image processing software, I'd be remiss not to mention Adobe Illustrator. EVERY scientist, in my opinion, should be familiar with at least the basic use of Illustrator for making publication-quality figures from raw images. This is especially true for anyone using Matlab. Every figure on the media/scientific-visualizations page on this site went through Illustrator, and I've never published a research article without Illustrator-polished figures. Note that this is not to distort or alter the image substantively, but to clean it up to make the data or effect or result easier for your reader to appreciate. I cannot overstate, though, that you never “doctor” your images or manipulate them in a way that fundamentally changes the substantive interpretation — post-processing should facilitate interpretation, not produce it.